Better Genome Editing
A major obstacle to in-cell genome editing is, well, the cell itself.
“Human cells don’t like to take in stuff,” explained UC Santa Barbara’s Norbert Reich, a professor in the Department of Chemistry and Biochemistry. The human cell has evolved a “trash disposal” mechanism that isolates and breaks down foreign proteins and other unwanted biomolecules, pathogens and even damaged cellular structures, he explained. So, for people in fields such as biotechnology, biopharmacology and genomic research and therapeutics — such as those working with the gene editing juggernaut CRISPR-Cas9 technology — results are only as good as their ability to efficiently bypass this defense mechanism and accurately introduce proteins into animal cells.
Reich and his team have developed such a method. Their technique, estimated to be 100 to 1,000 times more efficient than current methods, gives users complete spatiotemporal control of the genome editing delivery, in effect allowing them to decide exactly when and where to release genome editing proteins.
“We can actually hit individual cells,” Reich said. “We can even hit parts of a cell so we could release the protein into only a part of the cell. But the main point is that we have the control over where and when this protein that’s going to cut the DNA is going to be released.”
The research by Reich’s group, “Light-Triggered Genome Editing: Cre Recombinase Mediated Gene Editing with Near-Infrared Light” appears in the journal Small.
One attention-grabbing recent breakthrough in biotechnology is the use of gene editing proteins — “molecular scissors” such as CRISPR, Cas and, in this study, Cre — to find, cut and paste specific sections of target DNA sequences. Originally a defense mechanism used by bacteria and archaea to recognize DNA from attacking viruses and mark them for destruction, scientists have since developed methods of recognizing, cutting and binding base pair sequences of various lengths, using various proteins. The potential for this technology is massive, and ranges from basic research that determines the function and identification of genes to therapies that could fix cellular-level defects.
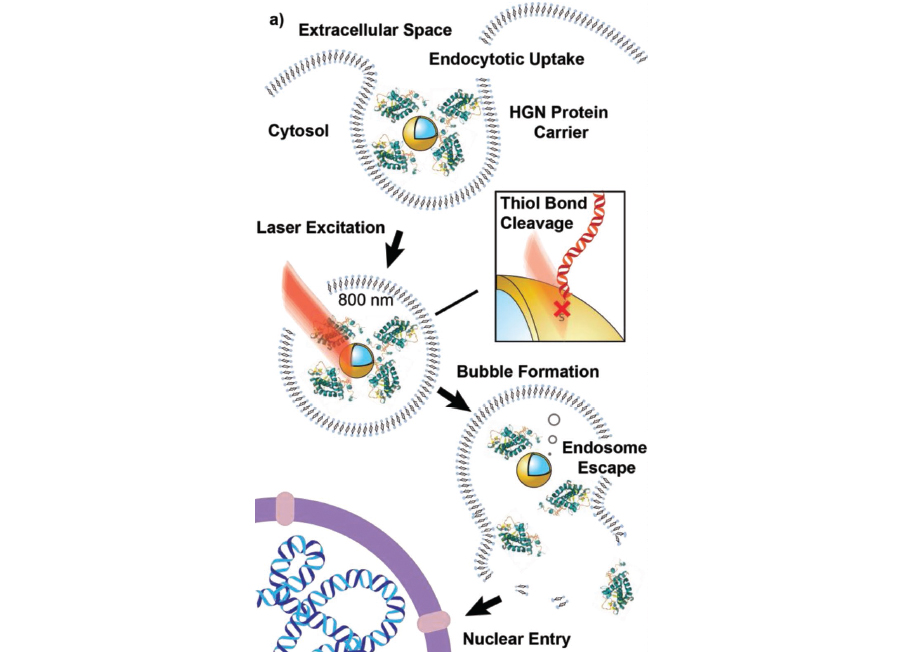
Key to the Reich group’s light-triggered genome editing are hollow gold nanospheres onto which are coated DNA reporter strands (they fluoresce red) and a protein fusion of Cre recombinase and cell-penetrating peptides. And near-infrared light.
“So now we’ve got a homing device and a delivery agent,” Reich said, explaining that the Cre recombinase and peptide fusion act as the targeting system, one that goes into play when the target cell does its cellular trash disposal.
Once taken into the cell, the nanoshell is enveloped in an endosome — a membranous pocket that isolates it and transports it through the cell.
“But the nanoshells don’t do anything because they’re entrapped,” Reich said. Ultrafast pulsed near-infrared laser light — which is harmless to cells and is efficient at tissue penetration — is then aimed at the entrapped nanoshells and their protein coats.
“Near-infrared wavelengths cause a really interesting thing to happen,” Reich said. “It causes the gold nanoshell to get excited and it causes whatever we’ve attached to come off.” At the same time, nanobubbles form, causing openings in the endosome and allowing its protein contents to escape. The proteins are now free to home in on the cell’s nucleus, where its genetic material is stored, and gain entry with the cell-penetrating peptide. And Cre can get to work finding, cutting and pasting its reporter strands into the helix.
The group’s in-vitro experiment proved successful: After a period of incubation, cells penetrated by the protein-coated nanoshells, followed by irradiation, glowed red.
“We didn’t engineer anything that would make the cells behave differently,” Reich said. “We made it so the cell would look different because of this fluorescent protein.”
Said the paper’s lead author, Dean Morales, who is now a postdoctoral researcher at Los Alamos National Laboratory, “As a basic research tool, with spatiotemporal control each cell can become an experiment. Imagine you’d like to study the function of a certain gene and how it alters that cell’s behavior or its behavior with a close neighbor. Using the plasmonic nanoparticles as an antenna we can either turn on or turn off a gene of interest and observe in real-time the ramifications of its activity.”
Spatiotemporal control also allows those who employ it to tread lightly on DNA, the rewriting of which, the researchers acknowledge, has very powerful and transgenerational effects.
“In certain cases, like somatic mutations, not every cell in the body would require editing,” Morales said. “The ability to control where and when the editing machinery can be used provides transience to the procedure. The importance of this is that current approaches to gene editing often result in the editing machinery being left in an active form in the targeted cell, with unknown long-term ramifications. Our approach delivers the editing machinery in a transient fashion, and thus circumvents this problem.”
Research on this project was conducted also by co-authors Erin Morgan, Megan McAdams and Amanda B. Chron of UC Santa Barbara; and Jeong Eun Shin and Joseph Zasadzinski, of University of Minnesota.