New Study Reveals Gene Expression Networks Underlying Age-Related Macular Degeneration

Age-related macular degeneration (AMD) is one of the leading causes of blindness worldwide, especially in developed countries. There is currently no known cure or treatment for the vast majority of AMD patients.
A new study led by scientists at UC Santa Barbara has identified genes whose expression levels can identify people with AMD, as well as genes that distinguish clinical AMD subtypes. The findings, which appear in BioMed Central's journal Genome Medicine, could offer new candidate targets for the development of AMD diagnostics and therapies.
It is estimated that 6.5 percent of people over age 40 in the U.S. have AMD. While smoking and UV light exposure are both risk factors for the disease, most of the risk for AMD is heritable. Several genes, including those involved in the innate immune system and fat metabolism, have been associated with altered risk for AMD. However, a detailed molecular description of the disease has yet to emerge.
"In large part, previous studies of AMD have taken a reductionist approach, focusing on one or a handful of genes or gene products at a time," said Monte Radeke, research scientist with UCSB's Neuroscience Research Institute and one of the project leaders. "We have taken a more holistic approach, directed at characterizing the entire scope of changes associated with AMD. By coupling this with a bioinformatic-driven analysis we can assemble a detailed, testable model that explains AMD."
Researchers at UCSB, the University of Utah's John Moran Eye Center, and the University of Iowa combined forces to tackle the issue. Tissue samples from a human donor eye repository were used to identify genes up-regulated in AMD. The ability of these genes to recognize AMD was tested on a separate set of samples obtained from the Lions Eye Bank of Oregon.
The team led by UCSB scientists discovered hundreds of genes with altered levels in AMD, the top 20 of which were able to predict a clinical AMD diagnosis. Genes over-expressed in the RPE-choroid (a tissue complex located beneath the retina) include components of cell-mediated inflammatory responses; while in the retina, the researchers found genes that function in wound healing and the complement cascade, a part of the innate immune system previously linked to AMD.
In donors with advanced stages of AMD, the researchers found extensive gene networks associated with neovascular AMD (in which the growth of new blood vessels obliterates the retina) and geographic atrophy (where the photoreceptors and RPE waste away). Consistent with the loss of vision in these advanced AMD stages, they found a decreased expression of a number of genes responsible for the detection of light.
Radeke explained: "Not only are these genes able to identify people with clinically recognized AMD and distinguish between different advanced types, some of these genes appear to be associated with pre-clinical stages of AMD. This suggests that they may be involved in key processes that drive the disease. Now that we know the identity and function of many of the genes involved in the disease, we can start to look among them to develop new diagnostic methods, and for new targets for the development of treatments for all forms of AMD."
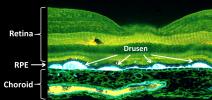
† Top image: Fluorescent micrograph of a macula with extensive drusen accumulation. Drusen accumulate between the RPE and the choroid. In addition to being sites of inflammation, drusen are believed to disrupt normal RPE and choroid function, leading to changes in gene expression and eventual loss of vision.
Credit: Courtesy of Dr. Robert Avery, California Retina Consultants.
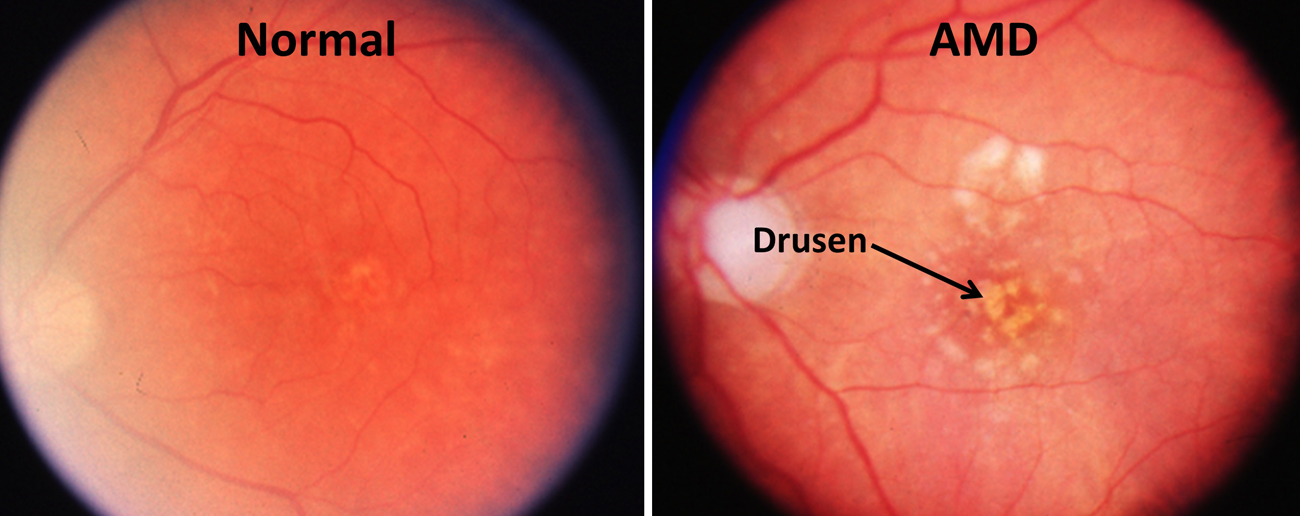
†† Middle image: Fundus photographs of the macular region of an eye from a normal individual and an eye from a patient with the dry form of age-related macular degeneration. The accumulation of sub-retinal deposits called drusen are evident in the AMD eye. Drusen, which appear as yellowish blotches in these photographs, are composed of lipids and proteins and are often sites of local inflammatory responses.
Credit: Courtesy of Dr. Robert Avery, California Retina Consultants.
Related Links



