Gut Ecology
For something that has evolved with us over millions of years, and remains part of our physiology over our entire lives, our gut microbiome, oddly, remains somewhat of a mystery. Comprised of trillions of microbes of at least a thousand different species, this community of bacteria, viruses, protozoa and fungi in our gastrointestinal tracts is unique to each individual and has been found to be intimately connected to various fundamental aspects of our fitness, from our immunity to our metabolism and mental health.
For UC Santa Barbara researchers Eric Jones, Zipeng Wang and Joshua Mueller, the gut microbiome is a remarkable machine, full of interactions and competitions that directly impact our wellness. Tuning this machine in a favorable direction, they say, could improve our resistance to disease.
“Can medical therapies improve host health by modifying the microbiome composition? We still don’t really know, so it’s a huge field of research,” Jones said. “This question is a great motivator.”
In our daily lives, many of us hope to improve our gut microbiome by taking probiotics and eating fermented foods. In clinical situations, fecal transplants have been shown to successfully treat recurring infections of the gut bacteria Clostridium difficile, which often recur after antibiotics used to treat infections wipe out “helpful” gut bacteria as well.
But outside of sweeping and heroic measures to add populations of good bacteria to the GI tract, there isn’t a whole lot known about how to subtly point the system in a healthy direction.
“It’s about more than just putting in the right microbes,” Jones said. “You need to understand the environment that the microbes are in and you need to understand what facilitates a stable gut microbiome composition.”
To tackle this problem, the researchers, under the guidance of UC Santa Barbara physics professor Jean Carlson, propose a technique for driving a mathematical gut microbiome model toward a target microbiome composition by manipulating certain parameters of the model. Called SPARC (SSR-guided parameter change), this approach reduces the complexity of the system without sacrificing it. And, according to a study published in the journal Physical Review E, it also “offers a systematic understanding of how environmental factors and species-species interactions can be manipulated to control ecological outcomes.”
Old equation, new use
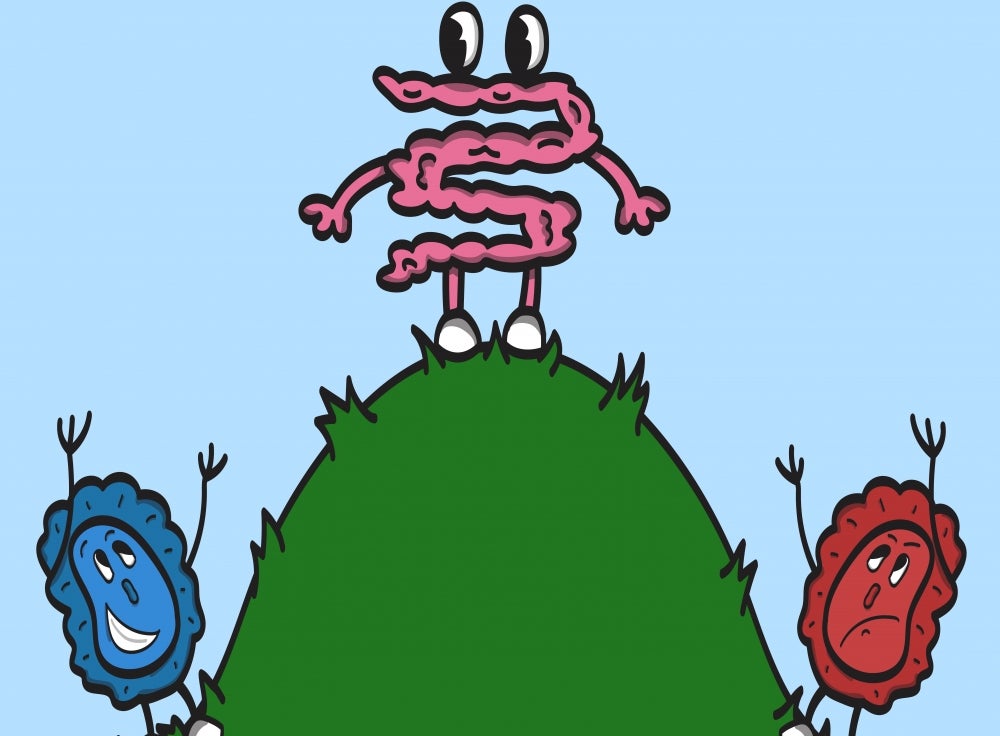
Photo Credit: DYLAN MUELLER ILLUSTRATION
“So basically, the goal is to find a parameter change that corresponds to a change in the microbiome environment,” said lead author Zipeng Wang. It helps to envision the gut microbiome as a ball perched on the top of a hill, poised to roll down in one direction or another. In this idealization, the gut environment dictates the shape of the hill. A healthy microbiome composition lies at the base of one side of the hill, and a disease-associated composition on the other. While fecal transplants directly push the ball to the healthy side of the hill, SPARC, according to the researchers, controls the shape of the hill, effectively rolling the ball down on one side or the other.
To describe this gut ecosystem (data was collected from mouse model experiments at Memorial Sloan-Kettering Center in New York), the researchers used the generalized Lotka-Volterra (gLV) equations. Known also as predator-prey equations, the gLV equations stem from a century-old method used in traditional ecology to describe the interactions between species on Earth, such as competition and predation, as well as effects from indirect interactions.
“But, one of the difficulties is that the gut microbiome has all these different bacterial species,” Wang said, “So the Lotka-Volterra equations become very high-dimensional, which means that there are a lot of parameters, and a lot of different ways for bacteria to interact with one another. It would be very hard for us to find the right parameters to achieve the desired microbiome composition.”
To avoid the trial-and-error of manipulating an unwieldy number of different parameters, the team opted instead to examine a compressed but reliable 2D representation of the ecological model generated by the dimensionality reduction technique called “steady state reduction” (SSR). According to the researchers, this allowed them to zoom out and identify the key parameters that control the shape of the hill.
“What we like about our model is that it gives us a systematic strategy to identify these low-dimensional features that are really sensitive, that are the most important,” Jones explained. “I was really surprised and pleased that we could, for example, find a single parameter and change it by 10% of its value and that would change the shape of the hill.”
What is that parameter? Well, given the diversity of gut microbiomes, diets, co-occurring conditions and environmental influences, there is not necessarily one universal parameter change — say, acidity, or fiber content — that fits all. The SPARC method, the researchers say, guides thinking on how to identify the significant parameters based on data.
In addition, SPARC currently is primarily a mathematical exercise, though the researchers are eager to try it out in an experimental setting.
“There are people working on what they call gut-on-a-chip systems, which are kind of like miniature Petri dishes that replicate some of the conditions of the human gut microbiome,” said Joshua Mueller. “It would be really exciting to validate SPARC in these tightly controlled experimental circumstances.”
In the rather more distant future, Jones said, this method could also help pave the way for personalized microbiome management, in which real-time readouts of the state of our microbiomes — say, from a smart toilet — could tell us to change our dietary habits to avoid illness and improve our general well-being.
“In order to get to that point, we need mechanistic models of the microbiome,” he said. “We need to understand how to control it. We need to understand how environmental feedbacks play into microbial dynamics.”



