
A new study suggests that protein knots, a structure whose formation remains a mystery, may have specific functional advantages that depend on the nature of the protein's architecture.
"The presence of a knotted or slipknotted structure in a protein is relatively rare but really is very interesting," said Kenneth Millett, a professor of mathematics at UC Santa Barbara and a co-author of the paper, "Conservation of complex knotting and slipknotting patterns in proteins," published in the Proceedings of the National Academy of Sciences.
Relatively little is known about protein folding, the process by which a polypeptide chain with a specific sequence of amino acid chains forms the three-dimensional structures –– their "native states" –– required to become functional. How this process reproducibly achieves the required structure is the subject of intensive study. Even harder is understanding how this is accomplished for knotted proteins, where the chain loops around itself in entanglements of varying complexity; or the even rarer slipknotted proteins, where a loop is bound by another segment of the protein chain, similar to a shoelace bow.
What intrigued the scientists about the protein knots is that the folding process resulting in the formation of knots is intrinsically more difficult than the process producing unknotted proteins. The protein has to avoid not only energetic traps but also topological barriers. If an amino acid chain takes too much time to find its native state or if it is stuck in a misfolded or partially unfolded state, the result may be a useless protein or one that produces harm by causing protein aggregation which is known to cause neurodegenerative disorders.
"From an evolutionary point of view, knotting might seem unlikely to occur but, in fact, it does occur," said Millett, who, along with co-first authors Joanna Sulkowska from UC San Diego and Eric J. Rawdon from the University of Saint Thomas and with Jose N. Onuchic from Rice University and Andrzej Stasiak from the University of Lausanne, examined, analyzed, and classified 74,223 protein structures submitted to the Protein Data Bank for the location and formation of knots. Millett worked on the development of the mathematical theory and the computer implementation needed to identify the location and type of knots in the proteins studied in the paper.
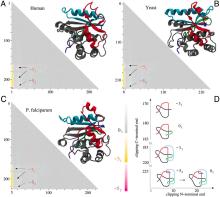
What they found was that protein knots and slipknots, instead of being discarded through the process of evolution, are often strongly conserved. This suggests that, despite their reduced efficiency of folding, the knots are somehow advantageous and important to the function of the protein.
Additionally, the researchers found that the location of these knots and slipknots is highly conserved, marked by points of flexibility –– "hinges" –– in the chain that may have properties necessary for more efficient folding.
Knots and slipknots could contribute to the stability of the protein, as shown by the similar slipknot loops observed in several families of proteins that form transmembrane channels –– the ducts through membranes of a cell, that allow certain materials to pass through. The slipknot, according to the authors, seems to strap together several transmembrane structures giving stability and forming the channel needed to allow passage through cell membranes.
The researchers will continue their study of the little-understood process of protein folding and knotting, said Millett.
"These knots may help to identify features that turn out to be important, and aspects of the structure that are more generalizable. We need to clearly understand how these things come to be, what are the implications of their structure, and how might one be able to somehow guide them," said Millett.
† Bottom image: Molecular structures and matrix presentation for ubiquitin C-terminal hydrolases from (A) human, (B) yeast, and (C) P. falciparum plasmodium cells form the same knotting motif.



